Scientists from the Hebrew University of Jerusalem say they have developed a groundbreaking technique that can help uncover hidden differences between seemingly identical bacteria and may open new avenues to cure bacterial diseases.
“Our method, Microcolony-seq, is important because it enables us to characterize the different subgroups of bacteria that are causing infection,” Dr. Raya Faigenbaum-Romm, a postdoctoral researcher, recently told The Times of Israel.
Examining diverse microcolonies of bacteria taken from a human infection with this new tool is giving scientists a better understanding of why antibiotics and vaccines sometimes fail. The discovery may point the way toward more precise treatments.
“Subpopulations in bacteria taken from a human infection keep their distinct behaviors for about ten hours of growth in the lab,” said Faigenbaum-Romm. “This means that, in our understanding, the different states they had inside the patient remain stable enough for us to study outside the body, which is especially interesting and surprising.”
The research, published in the peer-reviewed journal Cell, was led by Faigenbaum-Romm under the supervision of Prof. Nathalie Q. Balaban of the Racah Institute of Physics, together with Hebrew University professors Ilan Rosenshine, from the Faculty of Medicine, and Maskit Bar-Meir, from the Faculty of Medicine and Shaare Zedek Medical Center.
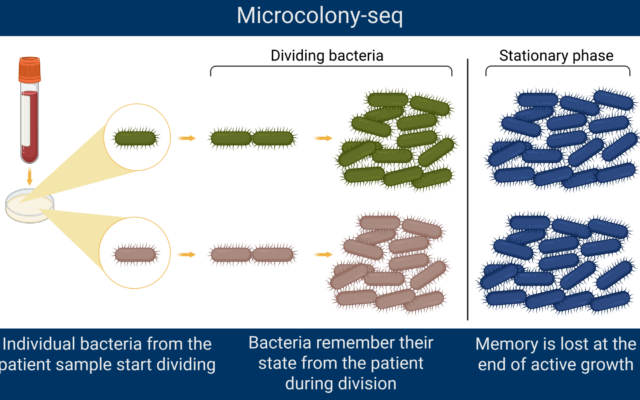
The Microcolony-seq method
Bacteria are invisible to the naked eye. To study microscopic organisms, scientists grow bacterial cultures in a controlled laboratory setting.
Individual bacteria divide very fast, explained Faigenbaum-Romm. In the microbiology lab, the scientists grow them on agar, which is “like a jelly with nutrients.”
Agar provides a stable surface that allows bacteria to grow into distinct clusters called colonies.
The Microcolony-seq method then isolates the tiny colonies, enabling researchers to gather material from many genetically identical cells that all originated from the same single bacterium. Their approach gives a clearer picture of coexisting subpopulations rather than analyzing single bacterial cells, even with cutting-edge single cell RNA sequencing methods. It also makes it possible to identify genetic differences between colonies.
Scientists can then understand why two cells might look or act differently.
At times, there are genetic mutations. At other times, the cells remain the same but the genes are expressed differently – for example, one subpopulation might have a higher level of iron metabolism – without a change in their DNA sequence.
The researchers studied tiny bacterial colonies while they were in the rapid-growth phase, growing side by side on the same agar plate. This means all the colonies experienced the same exact environment.
The scientists were then able to analyze the colonies when they were still actively dividing. Using this method, the team found unexpected results.
Pathogens such as E. coli, which is found in the urinary tract or in the intestines, and Staphylococcus aureus, which is a bacterial strain that can cause fatal infections, were each shown to split into stable subpopulations.
Some lineages were activated to cling to host cells, while others switched on the signals in genes that favored motility, or movement, or survival in harsh conditions.
“In the lab, Microcolony-seq showed that urinary tract and bloodstream infections can contain coexisting subgroups of bacteria,” Faigenbaum-Romm said. “For the Staphylococcus aureus, which is a pathogen that can lead to a life-threatening blood infection, to sepsis or to death, we identified three subpopulations in the same patient, each with a different way of clinging to host cells.”
These different populations showed changes that were passed down to the next generation in what Faigenbaum-Romm called a memory.
Researchers were able to study these tiny microcolonies and learn about their previous state through their memories.
“Perhaps this memory allowed the bacteria to survive in different host environments,” Faigenbaum-Romm explained.
However, she said that when the bacteria had no more nutrients and reached the stationary phase, the memory was erased, effectively resetting the population so that they “all became the same again.”
Neutrophil and Methicillin-resistant Staphylococccus aureus bacteria. (Credit: Courtesy/National Institutes of Health)
Clinical usage
“An infection is rarely a uniform population of bacteria,” said Balaban. “It’s more like a coalition of different players, each with its own strengths. To design therapies that truly work, we need to understand — and target — all of them.”
When doctors run standard clinical tests, they usually look at just one bacterial colony after the bacteria had stopped growing.
If that colony doesn’t represent the whole population, doctors might prescribe a treatment that doesn’t work against the hidden subgroups.

Dr. Raya Faigenbaum-Romm, left, and Prof. Nathalie Q. Balaban of Hebrew University of Jerusalem. (Credit: Photo of Faigenbaum-Romm, Courtesy/Dr. Tamar Szoke; Balaban, Courtesy/Hebrew University of Jerusalem)
For example, urinary tract infections are highly recurrent infections, Faigenbaum-Romm explained. Although the patient is treated with antibiotics, the bacteria are still growing, sometimes even during treatment, because the drug “probably targets only one subpopulation, and perhaps these reoccurring infections are due to a different subpopulation that escaped treatment.”
That might explain why so many promising drugs and vaccines — especially against Staphylococcus aureus — have failed in human trials. They target only one subgroup of bacteria, leaving the others free to cause trouble.
“Our method helps us to look at the different subgroups that are causing the infection,” she said. “This discovery could change how doctors treat bacterial illnesses.”
Beyond fighting infections, Microcolony-seq could open up entirely new areas of research. By giving scientists a way to study how bacteria diversify, the method might also be applied to fungi, the gut microbiome — which are the trillions of microbes living inside the body — or even industrial fermentation processes that produce foods and medicines.
“We’ve been treating bacteria as if they’re all the same, but they are different,” Faigenbaum-Romm said. “The research opens up avenues for better treatment. This is my goal, not only to do basic research, but also to improve public health.”