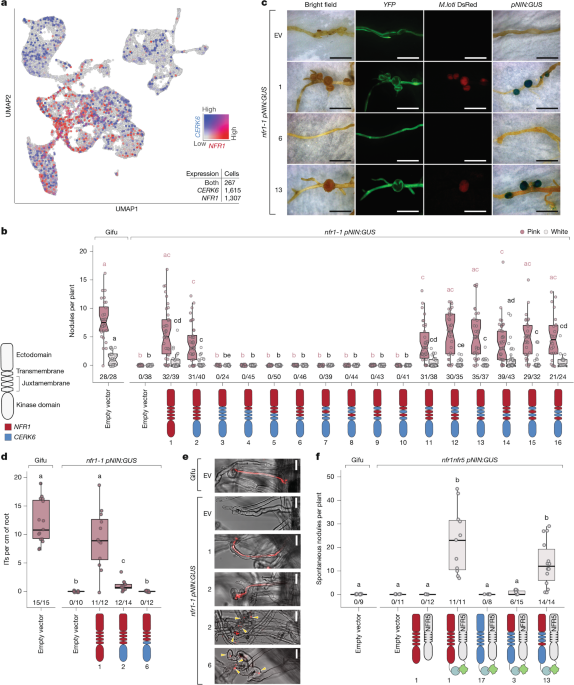
Bozsoki, Z. et al. Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science 369, 663–670 (2020). Article CAS PubMed ADS Google Scholar Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010). Article CAS PubMed PubMed Central Google Scholar Shiu, S. H. & Bleecker, A. B. Receptor-like kinases from Arabidopsisform a …
Read More »Tag Archives: Receptors
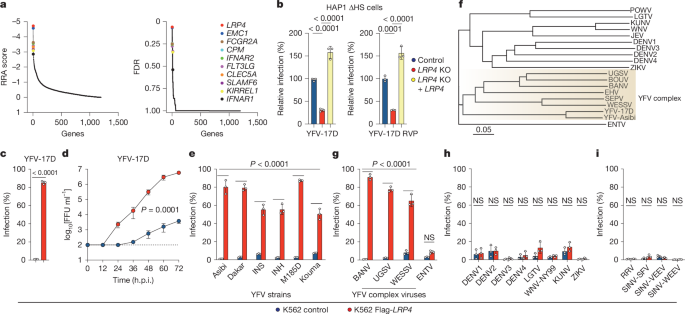
Multiple LDLR family members act as entry receptors for yellow fever virus
Pierson, T. C. & Diamond, M. S. The continued threat of emerging flaviviruses. Nat. Microbiol. 5, 796–812 (2020). Article PubMed PubMed Central Google Scholar Fishburn, A. T., Pham, O. H., Kenaston, M. W., Beesabathuni, N. S. & Shah, P. S. Let’s get physical: flavivirus-host protein-protein interactions in replication and pathogenesis. Front. Microbiol. 13, 847588 (2022). Article PubMed PubMed Central Google …
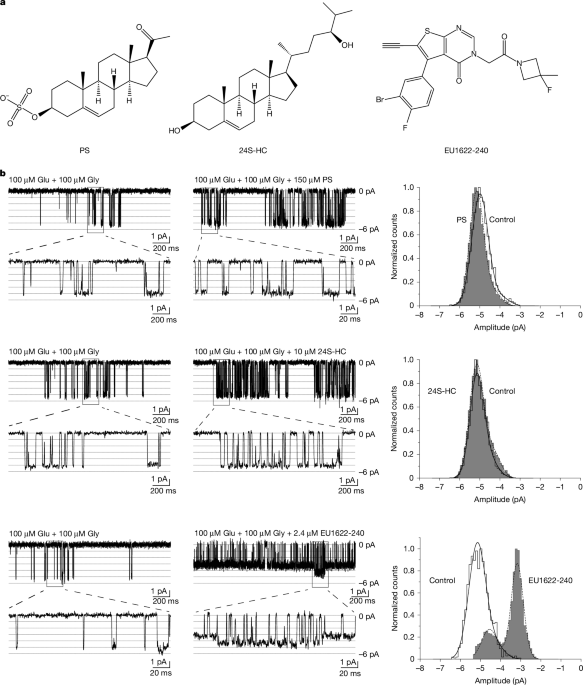
Read More »Mechanism of conductance control and neurosteroid binding in NMDA receptors
Hille, B. Ion Channels of Excitable Membranes 3rd edn (Sinauer, 2001). Jahr, C. E. & Stevens, C. F. Glutamate activates multiple single channel conductances in hippocampal neurons. Nature 325, 522–525 (1987). Article ADS PubMed Google Scholar Cull-Candy, S. G. & Usowicz, M. M. Multiple-conductance channels activated by excitatory amino acids in cerebellar neurons. Nature 325, 525–528 (1987). Article ADS PubMed …
Read More »AI Reveals Bitter Taste Receptors’ Hidden Role in Gut-Brain Health
Summary: Researchers used AlphaFold3, the latest AI-based protein modeling system, to predict the structures of all 25 known human bitter taste receptors (T2Rs). Compared with AlphaFold2, AF3 consistently generated more accurate structural predictions when benchmarked against experimentally determined data. The study revealed strong structural similarities in intracellular regions of T2Rs, with significant variation in extracellular regions, explaining how different receptors …
Read More »