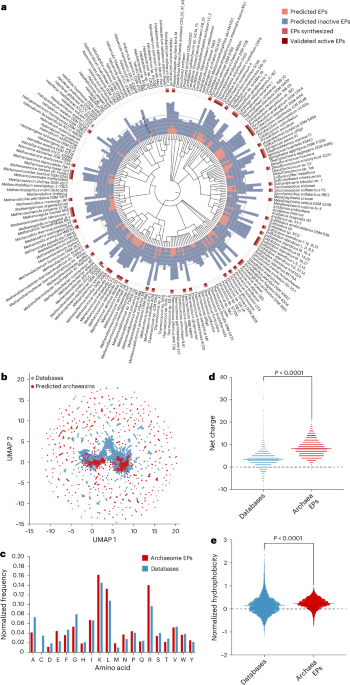
Deep-learning-guided identification of archaeasins We collected 18,677 non-redundant reviewed protein sequences from 233 archaeal organisms available on UniProt10 and used APEX 1.1, a deep learning antimicrobial activity predictor6 retrained on updated data (‘APEX 1.1’ in Methods), to mine EPs within archaeal proteomes. As APEX predicted bacterial-strain-specific minimum inhibitory concentrations (MICs), we used the mean MIC to represent the overall antimicrobial …
Read More »