Researchers showed that many mitochondrial proteins enter the organelle during synthesis, guided by folding patterns and structural signals. This discovery revises decades of biochemical models.
Mitochondria are organelles most commonly known as the “powerhouses of the cell” because they generate ATP (adenosine triphosphate), the main energy source for most cellular activities. They originated more than a billion years ago when an ancestral archaeal cell formed a symbiotic partnership with a bacterium.
Over evolutionary time, mitochondria became indispensable for metabolism and energy production, transferring most of their genetic material to the host cell. As a result, they now depend on the host to produce the majority of their proteins, which are synthesized by ribosomes outside the organelle and must be accurately transported into mitochondria.
Researchers at Caltech have now revealed new insights into how these proteins are moved from ribosomes in the cytosol, the fluid surrounding the nucleus, into mitochondria. Unexpectedly, they found that the pathway is strongly influenced by the mechanisms of protein folding.
“It turns out that localizing proteins to mitochondria involves a multilayered, complex pathway that is wired around the biophysical principles of protein folding,” says Shu-ou Shan, the Altair Professor of Chemistry at Caltech.
Challenging the traditional model
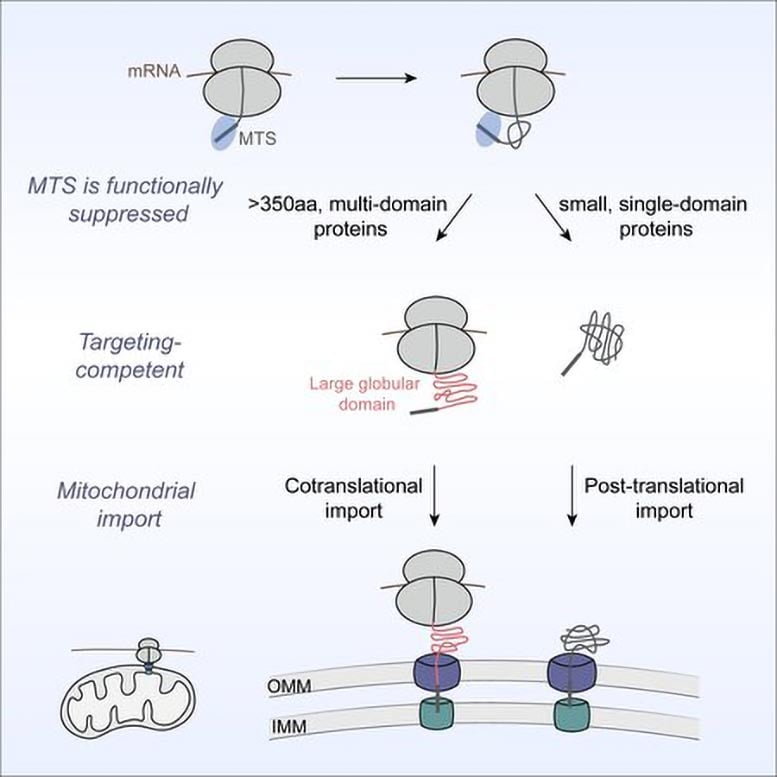
For many years, the prevailing view in biochemistry was that mitochondrial proteins are imported only after translation—the process in which ribosomes build proteins by linking amino acids together according to the genetic code—has fully finished. In a new study published in Cell, Shu-ou Shan and her team challenge this model, reporting that as many as 20 percent of mitochondrial proteins are cotranslationally imported. In other words, these proteins begin entering mitochondria while they are still being assembled on ribosomes.
“Once we identified these mitochondrial proteins that are cotranslationally imported, we asked, ‘What is special about this subset of proteins?’” says Zikun Zhu (PhD ’24), Shan’s former grad student and lead author of the paper.
Difficult-to-fold proteins and import timing
The researchers discovered that the defining trait of these proteins is their size and structural complexity. Many of them are topologically intricate, containing residues—amino acids within the chain—that may be far apart in sequence but must come together to achieve the correct three-dimensional fold. “That becomes a much more difficult process than just folding through interactions between neighboring residues,” Shan notes.
As a result, the system for cotranslational import into mitochondria prioritizes these really difficult-to-fold proteins. This makes sense if you consider that the large structures have to eventually go through narrow channels on the mitochondrial membrane during import. “There is going to be a problem if you let these large, very complex proteins finish translation in the cytosol,” says Shan. “They will get stuck in irreversible structures, and then you will not only block import, you will clog all the channels.”
Molecular signals and targeting sequences
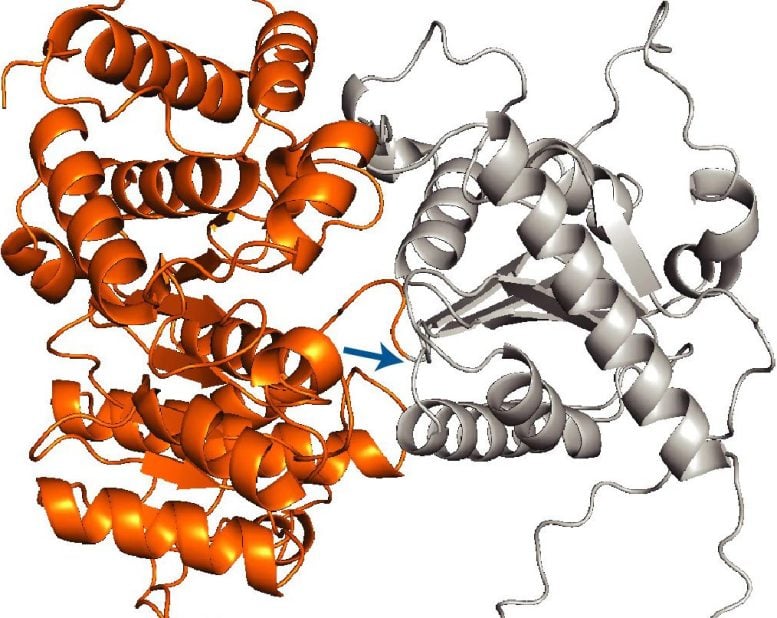
The team found that nearly all such proteins carry a mitochondrial targeting sequence, which is a signal that directs proteins to mitochondria. Yet, surprisingly, this alone is not enough to tell this subset of proteins to be delivered during translation. Zhu conducted experiments that showed that the system waits for a second molecular signal to move a protein to the mitochondria early. That signal comes in the form of the first large protein domain, or foldable structural unit within the sequence, that emerges from the ribosome.
“It’s like having your boarding pass locked in a suitcase,” Zhu says. “The targeting sequence is the boarding pass, but to access it, you need the code to open the suitcase. In this case, the large domain is that code.”
The scientists were even able to transplant examples of such large protein domains to other mitochondrial proteins that are normally imported after translation and showed that the domains indeed served as transferable signals capable of rerouting proteins to be imported during translation.
“Cotranslational targeting to mitochondria turns out to be completely different from targeting to other organelles,” Zhu says. “Going forward, it will be exciting to uncover more mechanistic details and, ultimately, to manipulate the timing of mitochondrial protein import. This will not only help us understand why cells evolved such a sophisticated targeting pathway for mitochondrial proteins but also open the door to potential therapeutic applications.”
Reference: “Principles of cotranslational mitochondrial protein import” by Zikun Zhu, Saurav Mallik, Taylor A. Stevens, Riming Huang, Emmanuel D. Levy and Shu-ou Shan, 11 August 2025, Cell.
DOI: 10.1016/j.cell.2025.07.021
This work was supported by the National Institutes of Health grant R35 GM136321 to S.-o.S. and the Howard Hughes Medical Institute through a Freeman Hrabowski Scholar grant to Rebecca Voorhees. E.D.L. acknowledges support from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 819318), by the Human Frontiers Science Program Organization (ref. RGP0016/2022), and by the Israel Science Foundation (grant no. 1452/18).
Never miss a breakthrough: Join the SciTechDaily newsletter.
Source link

